PNAS-2013-Gertz-E2772-81.pdf
Sirt3-MST-Nguyen 2013.pdf
GiangNguyen_PhDthesis_final.pdf
Sirt5 NAM inhibition_Fischer (PLOS one 2012).PDF
XG(12-16): Status of MicroCal VP-ITC.
- The MicroCal VP-ITC was delivered to PMC-AT lab on Dec. 5, 2013. It was then assembled and ready for test on Dec. 6, 2013. However, the computer did not start properly when power was on. XG and Joe Rivas had a brief phone conference and the problem was not solved. Joe offered to take the computer back and fix it.
- The computer unit was packed ready for shipping on Dec. 6, 2013.
- The computer unit was sent back to RT instrument on Dec. 9, 2013.
- It was received on Dec. 11, 2013.
Sherry & Risa (11-1 ): We are shopping for equipment which measures binding affinity of compounds (drugs). Above are comparisons of different analysis. Note the disadvantages. Sirtris Company uses SPR and ITC for their research. I will be shopping for SPR and ITC equipment because we got a green light to go ahead and buy.
| Current Interaction Analysis Tools |
Disadvantages |
||
| Surface Plasmon Resonance (SPR) |
This technique requires the immobilization of one binding partner on a sensor surface, raising issues of steric hindrance or molecular activity. These surface based methods are very sensitive in detecting minimal changes in the mass of sensor, but high affinity interactions are often not accessible for measure due to mass transport effects and transient binders are in most cases difficult to quantify. |
||
| Back-scattering interferometry (BSI) |
A label-free technology and do not need a label or surface coupling. It typically suffers from a refractive index background arising from non-reacted ligand, particularly at high concentrations. In addition, temperature-induced changes of the refractive index may interfere with the binding signal and render the interpretation difficult. |
||
| Isothermal titration calorimetry (ITC) |
A label-free technology and do not need a label or surface coupling. It can conduct measurements at a relatively low throughput and with exceptionally high sample consumption. |
||
| Fluorescence correlation spectroscopy (FCS) |
FCS relies on a significant change in the diffusion constant upon binding and is only applicable to a limited number of biomolecular interactions. |
XG (11-18): A summary of methods for measurement of protein-ligand binding affinity was attached. Summary of advantages and limitations of FP.ITC.SPR.FCS.FLIM.FRET.TIRFM.SPA.docx
RC (11-4): Have you find any published protocols on experimental on/off rate determination?
XG (11-18): The experimental on/off rate determination was barely described in publications. One called Competitive Radioligand Binding experiments. The kinetics of competitive radioligand binding predicted by the law of mass action_1984_Motulsky.pdf It is commonly performed with a single concentration of radioligand and a variety of concentrations of competitor in order to generate a competitive binding curve. One application of this model is attached. (Structure-kinetic relationships-An overlooked parameter in hit-to-lead optimization. Structure-kinetic relationships an overlooked parameter in hit-to-lead optimization_a case of cyclopentylamines as chemokine receptor 2 antagonists_2013_Vilums.pdf It does not quiet fit to our model. Will dig more if there is any protocols available.
RC (11-19): Which method is being discussed today?
XG (11-19): Microscale Thermophorosis (MST) is going to be discussed today. I do request the speaker to talk more about what makes MST stand out of the other available instruments, especially ITC, like MicroCal iTC200, MicroCal VP-ITC (the one we are currently interested in purchasing).
XG (11-19): Summary of MST seminar
- Compare with MicroCal ITC, MST (1) uses much less amount of sample, (2) buffer independent and purification free, and (3)quick readout (15 min per sample). In our case, (1) small molecules are our main interest and enzyme wil be made in the lab therefore sample amount will not be a critical issue. ITC will be fine. (2) ITC needs buffer optimization. For a target protein, the optimization will be one time deal. (3) As mentioned, ITC need more time for the instrument cooled down. However, it depends on the nature of protein, and may not require for high temperature for binding. In conclusion, the advantages MST claimed are attractive, but ITC is still good for our current applications.
XG (11-20): To estimate of the efficiency in the drug screening throughput between ITC and MST, we should consider the time spent on the following steps (1) Optimization; (2) Sample preparation; (3) Measurement; (4) Data analysis.
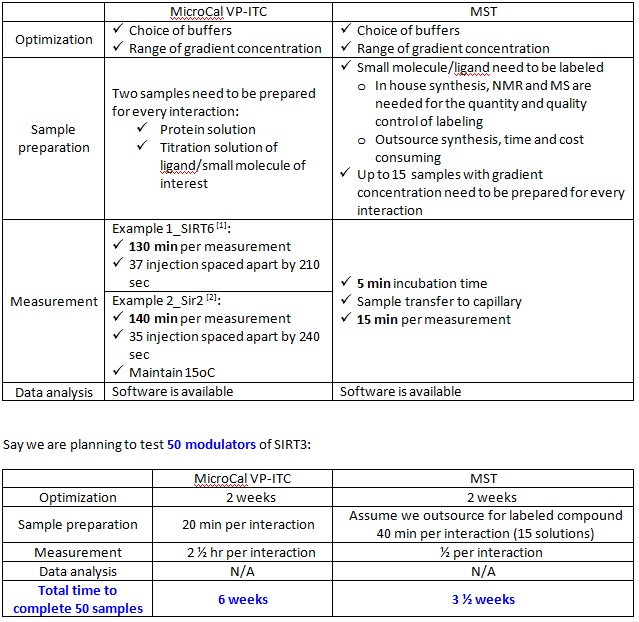
[1] J. Biol. Chem. (2011) 286: 14575-14587.
[2] J. Mol. Biol. (2004) 337: 731-741.
- Two available CRO in Germany (1) Crelux http://www.crelux.com/ (2) 2bind GmbH http://www.bioregio-regensburg.de/index2.php?mid=1214. The cost of outsource service is ~ $2000 per interaction.
- MST-Selected publications in 2013
- Sensitivity of MST and ITC: MST is fell in range of pM - mM and ITC is in the range of Sub uM-mM.
- Small molecule labeling will be extra work and cost. Issues like flourophore binds to protein need to be future addressed.
- Monolith NT.115 Series: ~ $130,000/ Monolith N.T. 119 Seies: $160,000 / Monolith NT.LabelFree: $190,000.
SM (7/28/16): Notes on labeling the protein for MST experiments - information obtained from Thomas Schubert of 2bind (Germany)
"We are using a NHS chemistry to label proteins. This means lysines are randomly labelled using this procedure. Statistically one label is attached per protein in a random manner. The labelling reaction is usually done in the protein storage buffer if possible. Please note, that Tris buffer have to be exchanged to other buffers (this is also done by us and is included in the costs). Tris contains primary amines, which would also be labelled by the NHS coupling, thus it has to be replaced by another buffer substance.
As the labelling is more or less random, the influence on the activity of the protein is usually rather low or not present.
Comparing the labelfree and the labelled assay type:
In your specific case of protein-ligand interaction, the labelled assay is in my eyes more adequate.
1. empirically every 4th compound shows background in the labelfree detection mode. These compounds can not be measured in the labelfree version. Only in the fluorescent MST binding assay.
2. the sensitivity of the fluorescent assay is better. Affinities as low as 10 pM can be seen in this assay type. In the labelfree assay the lowest affinity you can determine in a proper manner is around 50 nM (if the protein contains a lot of tryptophanes that are solvent oriented).
One option that I can offer to you is:
We can test the compound in the labelfree system for background. And we can test the protein for the specific fluorescence in the labelfree system. If both are fine, we can try the labelfree system. If not, we can switch to labelling of the protein and continue there. The necessary amount for both assays is more or less comparable. (Labelfree might be consuming a bit less)."